Computational Modeling of Ph Effect on ABE Fermentation using three Different Clostridium Acetobutylicum Strains Fed with Various mixtures of Glucose/Xylose
Project Details
- Student(s): Ibrahim Ghannam
- Advisor(s): Dr. Elie Chalhoub | CO- Contributor: Dr. Joanne Belovich, Professor, Chemical and Biomedical Engineering, Cleveland State University, Ohio, USA
- Department: Petroleum
- Academic Year(s): 2024-2025
Abstract
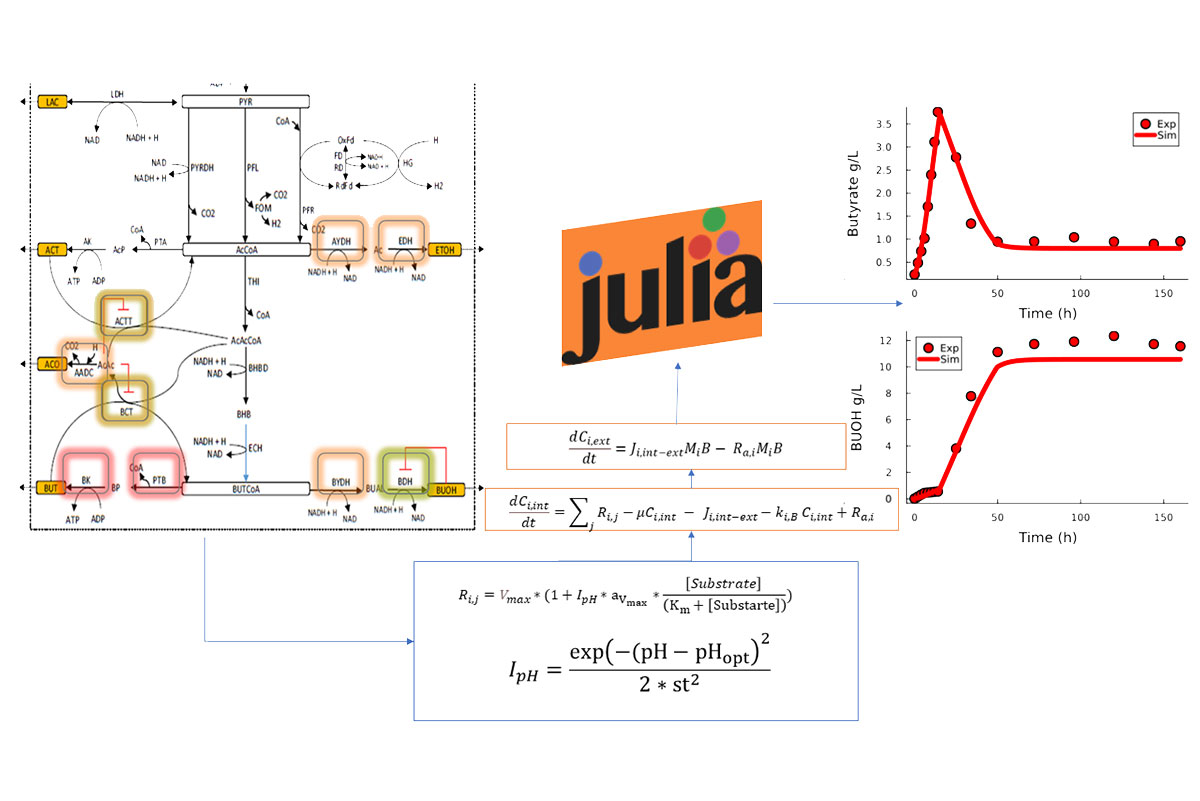
Acetone-butanol-ethanol (ABE) fermentation using Clostridium Acetobutylicum and lignocellulosic feedstocks is a renewable source for the production of bio-butanol. However, several challenges still face this biological route such as, finding optimum pH conditions, and effectively consuming the various mixtures of sugars present in lignocellulosic sources, hence affecting the yield and productivity of butanol product. There has been immense experimental research work dedicated to improve bio-butanol production process. On the other side mathematical modeling has proven to be an integral part in assisting experimental work by efficiently testing various hypotheses aiming at improving butanol production by saving operation cost and time. A detailed mechanistic model of ABE fermentation was developed in this work that incorporates the effects of various external pH conditions on cell growth and key metabolic enzymes. The model represents the fermentation of glucose under batch and continuous mode using three C. acetobutylicum strains (DSM 792, P1201, and YM1). The model was fitted and validated using various experimental data from published literature, and it was further analyzed using sensitivity analysis to assess the impact of key kinetic parameters on acid and solvent production as function of pH conditions. The model achieved from this study was found to be in good accordance with experimental data representing the effect of various pH conditions Our model was able to predict that the strain YM1 prefers an uncontrolled pH environment which led to a butanol production of 14g/L. While for the case of DSM 792 and P1201, the model was also able to mimic the strains higher butanol production in the case of controlled pH at 12hours. In addition, the model was also able to adhere to the pH neutralization using CaCO3, and buffering using NaOAc/HOAc.